Contact Advanced Research Computing
Quick Links
Current projects
Epidermal Growth Factor Receptor Analysis with GenFit: A Fitting Tool for Rule-based Models
Brandon Thomas, Biological Sciences
The epidermal growth factor receptor (EGFR; ErbB-1; HER1 in humans) is the cell-surface receptor for members of the epidermal growth factor family (EGF-family) of extracellular protein ligands (Herbst 2004). A number of cancers are driven by a mutation in the EGF receptor. Thus, having a thorough knowledge of the mechanism behind EGFR and its related components may help aid researchers in the search for effective cancer treatments. We’re using a new software, in conjunction with Monsoon, to run simulations modeling the EGFR signaling pathway. The software, GenFit, is a fitting tool for rule-based models. It employs a genetic algorithm while running thousands of simulations. These simulations “breed” with each other to produce progressively accurate models as the number of generations increases. Monsoon has already helped us produce results for the EGFR model, and more work is currently underway.
GenFit – A fitting tool for rule based models
Rule-based models are analyzed with specialized simulators, such as those provided by the BioNetGen and NFsim software packages. BioNetGen translates rule-based models into models having traditional forms, such as differential equations, and implements deterministic, stochastic and hybrid simulation methods. NFsim implements a network-free simulation algorithm, which is a particle-based kinetic Monte Carlo method in which rules are used as reaction-event generators. Network-free simulation allows one to cope with rule-based models that cannot be recast into traditional forms. Currently, when using a network-free simulator, fitting (i.e., optimizing parameter values for consistency with data) requires purpose-built scripts. The lack of general-purpose software tools for fitting has limited the use of rule-based models to analyze data, as well as the reproducibility of analyses involving fitting. GenFit is a general-purpose tool for fitting that is compatible with simulators for rule-based models, including BioNetGen and NFsim. GenFit is designed to take advantage of distributed computing resources. This feature enables fitting where simulation is computationally expensive, which is often the case when using a network-free simulator.
Asteroid Project
Michael Mommert, Physics and Astronomy
In his analysis, Mommert found that about two thirds of the volume of the asteroid consists of void space. This empty space is either of microscopic nature in the form of a very porous rocky material (left image), or it is a results of large sub-surface cracks and voids that are covered with a powder-like material called regolith, concealing their existence.
Chromosome Sequence of Burkholderia Pseudomallei
The Pathogen and Microbiome Institute (PMI)
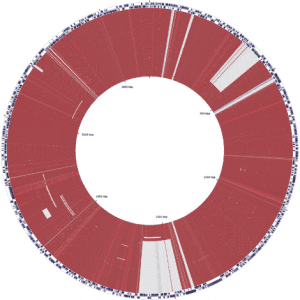
The fully sequenced chromosome 2 from more than 100 Burkholderia pseudomallei isolates from a single patient over 13 years is shown here. The isolates are chronologically ordered from oldest isolate-year 2000 (in the center) to the youngest isolate-year 2013 at the outer ring. This figure demonstrates that there is large scale gene shedding/loss over time, as evidenced by large deletions (large white blocks at about 1 o’clock and 6 o’clock and thinner white block at 10 o’clock) that occur part way through the infection (around year 2004 and 2013). These deletions are maintained throughout the rest of the infection (out though the outer ring/year 2013). In contrast, there are smaller, more intermittent deletions that occur in one or a few isolates (circles) but that are not maintained over time, as evidenced by smaller events. Gene loss is expected in long term infection, and this fundamental idea is true for B.pseudomallei.
Computational Cardiovascular Biomechanics
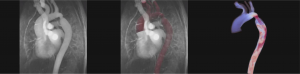
Amirhossein Arzani, Cardiovascular Biomechanics Lab
Our research group focuses on using computational tools to study the mechanisms behind initiation and progression of different cardiovascular diseases. Particular attention is given to developing computational models that can capture the multiscale and multiphysics nature of cardiovascular disease. Our work spans a variety of disciplines, such as Computational Fluid Dynamics (CFD), computational nonlinear structural mechanics, and mass transport. All of our simulations are done using open-source software that support High-Performance Computing.
Phenocam

PhenoCam pictures (winter, spring, summer, autumn) from Hart Prairie, located north of Flagstaff on the west side of the San Francisco Peaks.
The PhenoCam Network uses repeat digital photograph to track the rhythmic seasonality of vegetation activity in ecosystems across North America and around the world. This study of this seasonality is the science of phenology, and a phenocam is a digital camera specifically used to study phenology. From Alaska to Arizona, and from Hawaii to Maine, phenocams upload images to our server at NAU every 30 minutes, from sunrise to sunset, 365 days a year. Images and data products characterizing vegetation “greenness” are made publicly available in near-real time through the PhenoCam web page. The PhenoCam archive includes over 80 million images from almost 1000 different cameras. With a total of more than 6000 site-years of derived phenological data, and 75 cameras that have been in continuous operation for over a decade, the PhenoCam Network archive is the largest repository of phenologically-relevant repeat digital photography in the world. In addition to scientific research (over 200 papers have been published using PhenoCam Network data and imagery), PhenoCam provides unique opportunities for education and outreach. Engage in virtual travel through time and space by browsing the phenocam image archive!
GRIME-AI
Digital cameras are now widely used tools for environmental and ecological science, e.g. using camera traps to monitor wildlife, or PhenoCams to track vegetation seasonality. However, extracting scientifically useful data from images is not always easy. In a collaboration between hydrology researchers and software developers at the University of Nebraska-Lincoln, NAU’s ARC group, and Andrew Richardson’s PhenoCam team, Monsoon is being used as a test platform for an AI-based software platform called “GRIME-AI.” An overarching goal of the project is to lower the barriers to image processing and data extraction, allowing people with different levels of technical expertise and backgrounds to advance science using digital repeat photography.
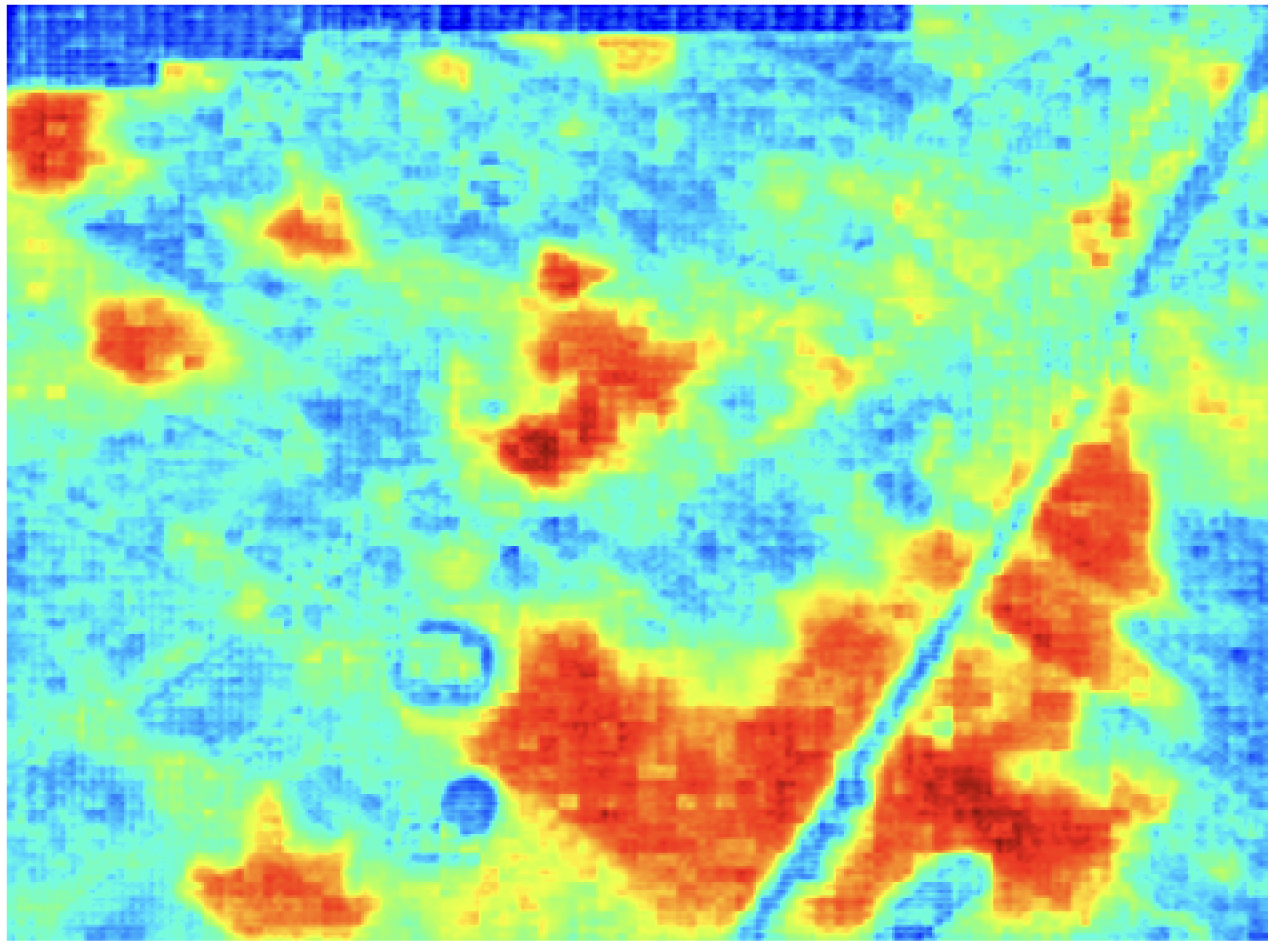
One application of GRIME-AI that is currently being worked on is the automated segmentation of different types of vegetation in tens of thousands of PhenoCam images from 72 cameras installed at a rainfall experiment in New Mexico. This is challenging because the amount, color, and texture of vegetation vary throughout the year. Some plots are predominantly bare soil, while others are mostly covered in vegetation. In addition, the plot infrastructure casts strong shadows in most images. The probability heatmap on the right shows how GRIME-AI classified each pixel in the image on the left, where red indicates a very high probability of grass and blue indicates a very low probability.